
VISUALIZATION OF MOLECULAR INTERACTIONS & PROPERTIES
Grapheme™ TK
2D structure depiction of molecules is the "natural language" of chemists, since this representation instantly conveys many important molecular properties.
Historically, 2D representations have mainly been used to visualize the connection table of molecular graphs. However, projecting 3D information into the 2D layout and displaying various atom and bond properties on the molecular diagram opens up a novel way to present information to chemists.
Grapheme TK provides several representation schemes that allow visualization of complex molecular interactions and properties in a clear and coherent 2D format that is the most natural to chemists.
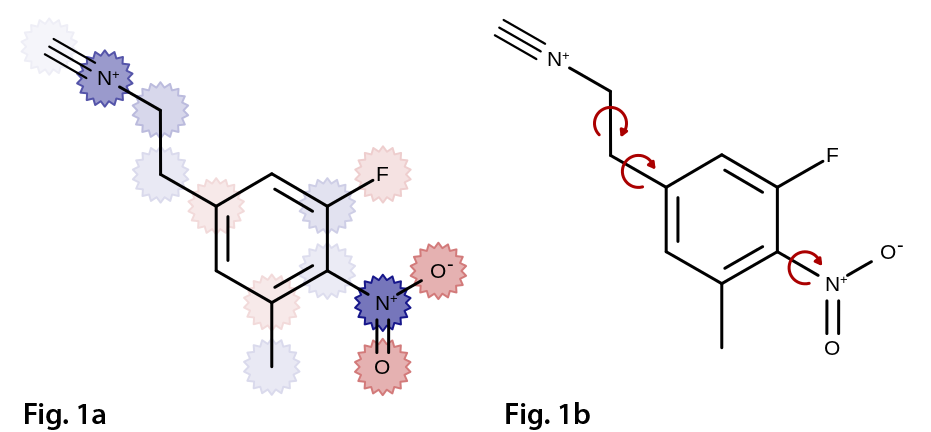
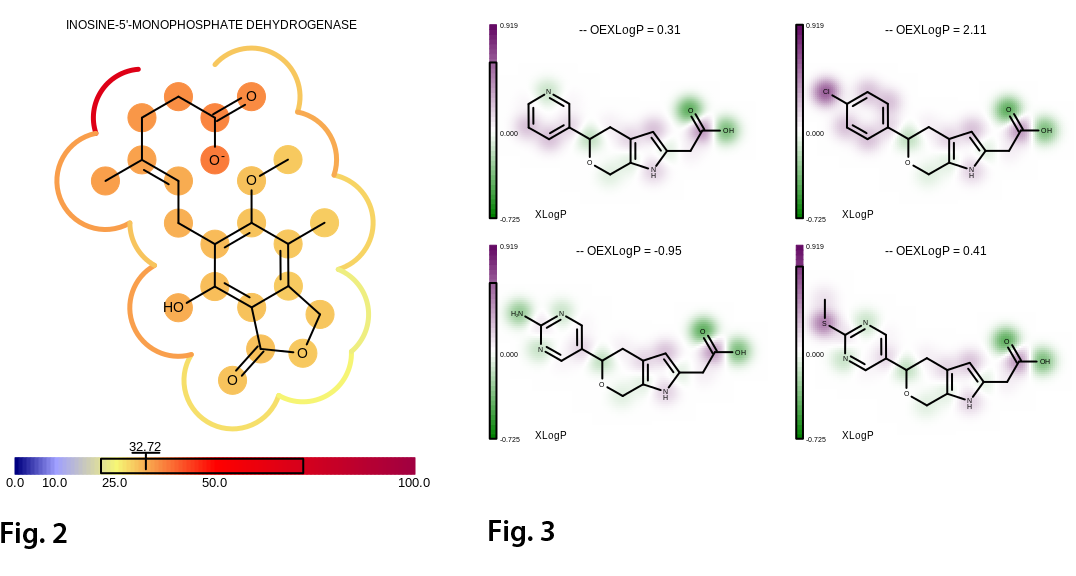
Features
- Wide range of built-in atom and bond glyphs for annotation
- Molecule surface
- Property map
- 2D coordinates generation driven by 3D conformation
- Visualizing ligand-protein complex surface
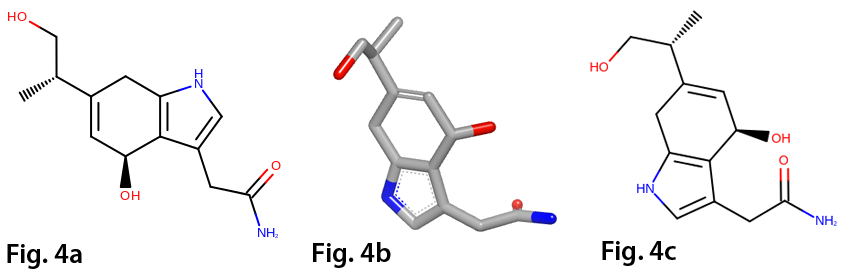
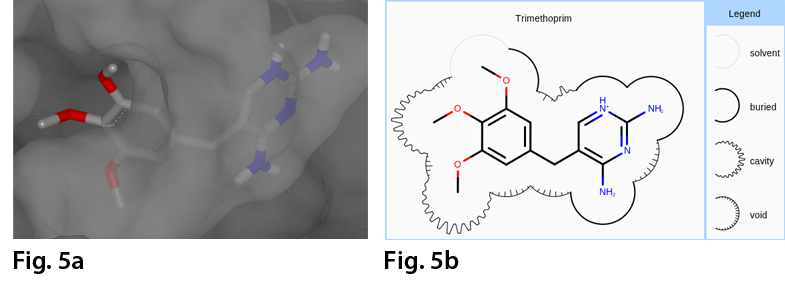
For more detailed information on Grapheme TK, check out the link below:
DocumentationCheminformatics
The Cheminformatics suite of toolkits provides the core foundation upon which all the OpenEye applications and remaining toolkits are built.
- OEChem TK Core chemistry handling and representation as well as molecule file I/O
- OEDepict TK 2D Molecule rendering and depiction
- Grapheme™ TK Advanced molecule rendering and report generation
- GraphSim TK 2D molecular similarity (e.g. fingerprints)
- Lexichem TK name-to-structure, structure-to-name, foreign language translation
- MolProp TK Molecular property calculation and filtering
- Quacpac TK Tautomer enumeration and charge assignment
- MedChem TK Matched molecular pair analysis, fragmentation utilities, and molecular complexity metrics
Modeling
The Modeling suite of toolkits provides the core functionality underlying OpenEye's defining principle that shape & electrostatics are the two fundamental descriptors determining intermolecular interactions. Many of the toolkits in the Modeling suite are directly associated with specific OpenEye applications and can, therefore, be used to create new or extend existing functionality associated with those applications.
- OEChem TK Core chemistry handling and representation as well as molecule file I/O
- FastROCS™ TK Real-time shape similarity for virtual screening, lead hopping & shape clustering
- OEDocking TK Molecular docking and scoring
- Omega TK Conformer generation
- Shape TK 3D shape description, optimization, and overlap
- SiteHopper TK Rapid Comparison of Protein Binding Sites
- Spicoli TK Surface generation, manipulation, and interrogation
- Spruce TK Protein preparation and modeling
- Szybki TK General purpose optimization with MMFF94
- Szmap TK Understanding water interactions in a binding site
- Zap TK Calculate Poisson-Boltzmann electrostatic potentials
miniWebinar: Opening up the druggable universe one cryptic pocket at a time
miniCUP Cambridge 2025
Expanding Orion’s Capabilities with AI
miniWebinar: Faster, Larger, Smarter: Filling the Funnel for Ultra-Large Scale Virtual Screening
Resources
Glimpse the Future through News, Events, Webinars and more
Event
miniCUP Basel 2025
Webinar
miniWebinar: Opening up the druggable universe one cryptic pocket at a time


