Just recently, Swann et al. of Abbott Labs published "A Unified, Probabilistic Framework for Structure- and Ligand-Based Virtual Screening" in the Journal of Medicinal Chemistry. If you haven’t read it yet, I highly recommend it. The paper is a very interesting extension of previous work done by Muchmore et al., also of Abbott Labs. Muchmore’s paper, "Application of Belief Theory to Similarity Data Fusion for Use in Analog Searching and Lead Hopping," presented a system for calculating a quantitative estimate of the likelihood that any two molecules will exhibit similar biological activity based on ligand similarity. Swann’s paper extends this work to include information obtained from structure-based virtual screening using docking.
He focuses primarily on three metrics in the paper: the CGO score from the FRED docking program, the combined shape and color Tanimoto scores from ROCS, and the 2D fingerprint similarity calculated using ECFP6. This is remarkable because they have created a unified and extensible system that effectively combines both structure- and ligand-based information in a meaningful way to assign probabilities of equipotency between any two molecules. While I’m on the subject of the utility of combining structure- and ligand-based information, I’d like to recommend another interesting paper, "FRED Pose Prediction and Virtual Screening Accuracy" by Mark McGann of OpenEye, also published in February, which shows that there are real statistically significant benefits to combining both these types of information when performing virtual screening using the Hybrid Docking functionality of FRED combined with ROCS results.
Getting back to the Swann’s paper, what I find particularly exciting is that this system can be recreated entirely using OpenEye software. Clearly, both FRED and ROCS are part of the core OpenEye suite, but what people might not know is that the upcoming 2.0 release of GraphSim TK will feature two new fingerprint types (circular and tree) that have been shown to be equivalent in performance to the ECFP6 fingerprints used in the paper.
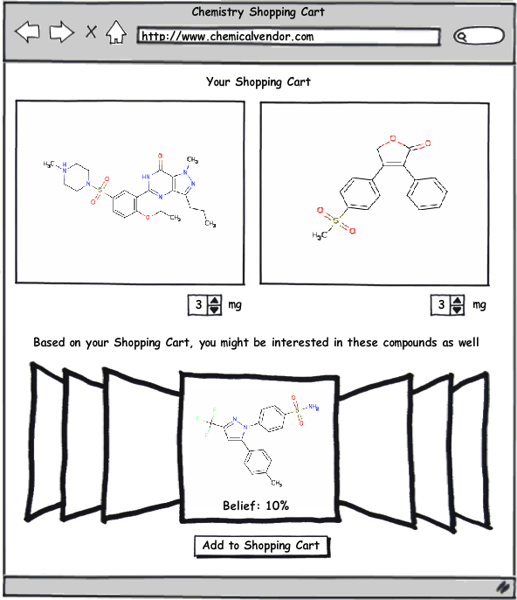
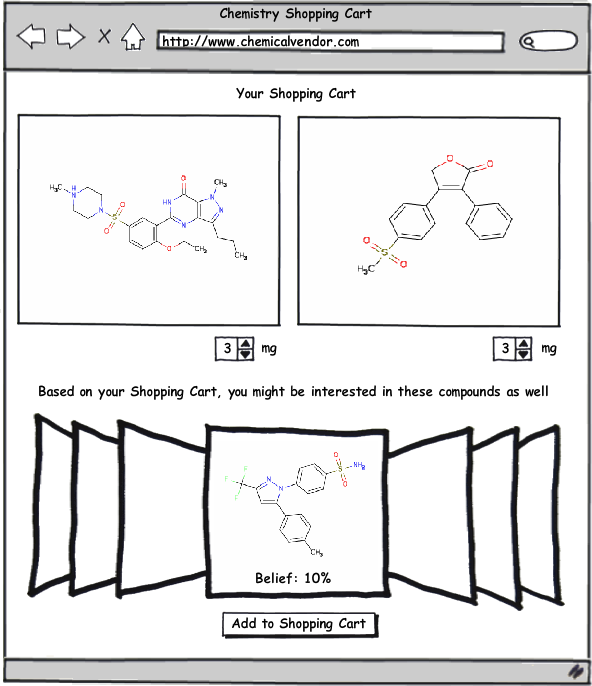
Obviously this system has many practical benefits to pharmaceutical and biotech companies, but another area where I think it could be readily applied is as the basis for a recommendation system (like those used by Amazon.com or Netflix) for compound selection by chemical vendors. Imagine for a moment going to a compound supplier’s website—and every time you add a compound to your shopping cart, a query is sent to a server that uses the techniques described in the papers by Swann and Muchmore to build a list of all the compounds available for purchase in their database that have a certain threshold probability of equipotency to those in your shopping cart. To make this system work, the software being utilized must be able to return results very quickly. Fortunately, this is an area where OpenEye software excels, particularly when you consider the enormous speed improvements available with FastROCS. I think this system could potentially be a very profitable tool for the compound vendors as well as a great service to those purchasing from them.
This recommendation system doesn’t have to be limited to compound vendors; it could just as easily be adapted to work internally with a company’s corporate collection (as well as other virtual libraries). A great example of this would be as a post-processing step after running an HTS screen. All the HTS hits would automatically be fed into this system, which would then recommend additional compounds based on the hits. This would broaden searches outside the compounds in the HTS screen as well as potentially recovering some false-negatives from the screen. One advantage of this approach is that it can take advantage of in-house structural information (as described in the Swann paper) to improve prediction accuracy over just the 2D and 3D methods that might be used by an external service.
I am very excited about the implications of the work done by Swann et al., and am looking forward to hearing more about the new and innovative ways people are using our tools to combine structural information with both 2D and 3D ligand information.