
Orion® MacroMolecular Data Service
Curated and Ready-to-use Protein Models
In drug discovery, quality of structural data is key to understanding modeling results. Preparing proteins from experimental data and predicted models for structure-based discovery contains many critical steps.[1] In the Orion MacroMolecular Data Service (MMDS), we have simplified the process for you.
With Orion MMDS, you have access to more than 100,000 pharmaceutically relevant protein structures from the Protein Data Bank (PDB) and AlphaFold, encompassing more than 16,000 targets. These proteins are readily prepared for your modeling needs, classified by functions,[2] and annotated for model quality.[3] You also can prepare and upload* protein structures from your own in-house collection.
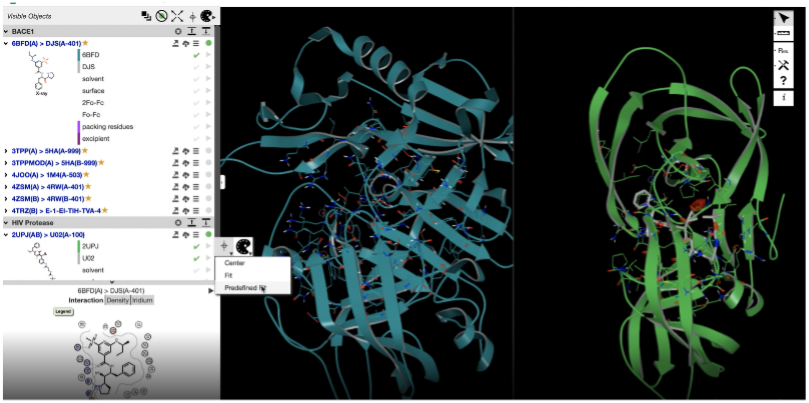
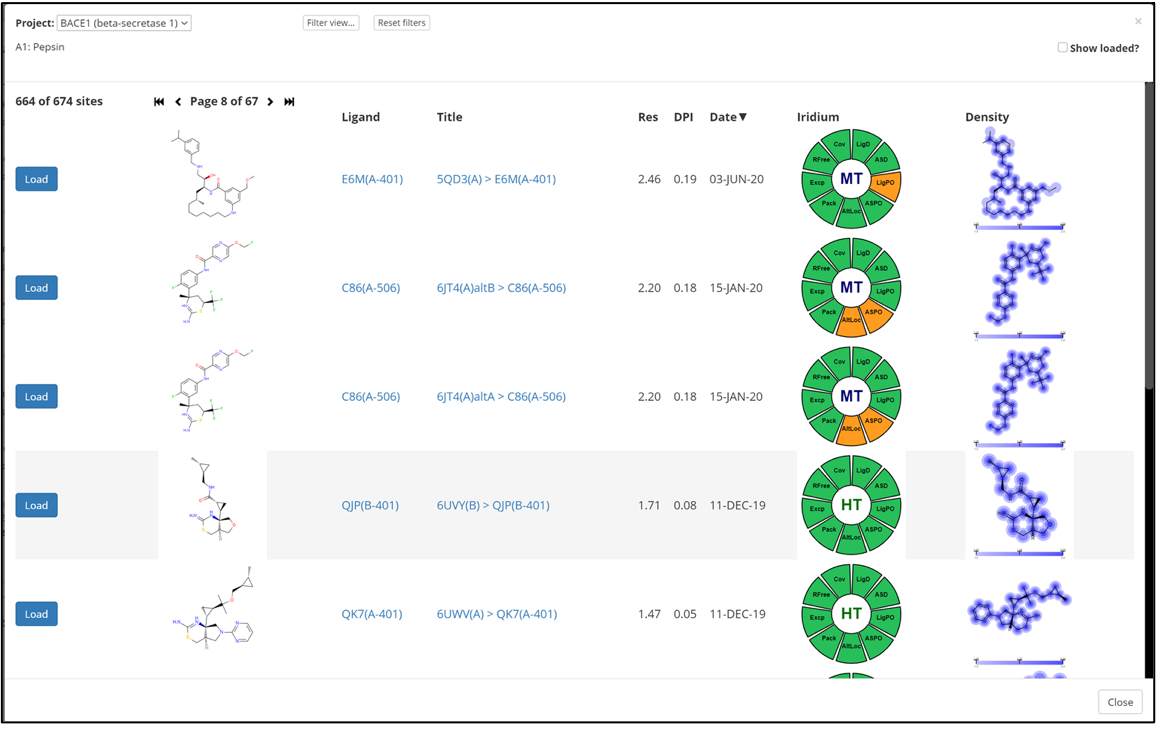
Fast Structure Loading and List Curation
Directly access >100,000 highly curated protein structures. Or load your structures directly using pre-built workflows in Orion*
- Input data from the PDB or from internal lists.
- Upload just the protein/target or include additional components, such as ligand and solvent.
- Tag data to identify experiments, sites, contexts.
- Arrange structures by family and sub-family.
- Curate and share structure lists with internal users.
- Easily add, remove, or rearrange structures within project lists.
- Use pre-built Orion Floes (guided workflows) or create your own custom workflow.
Reliable Scoring and Assessment
Prepare, score, and assess protein structures using trusted OpenEye science.
- Choose whether to analyze just your protein/target or include additional components, such as ligand and solvent.
- Assess model quality using Iridium criteria.[3]
- Integrate the proteins or ligands for use in OpenEye’s Gigadock™ functionality and through FastROCS shape.
- Perform similarity searching within the Orion platform.
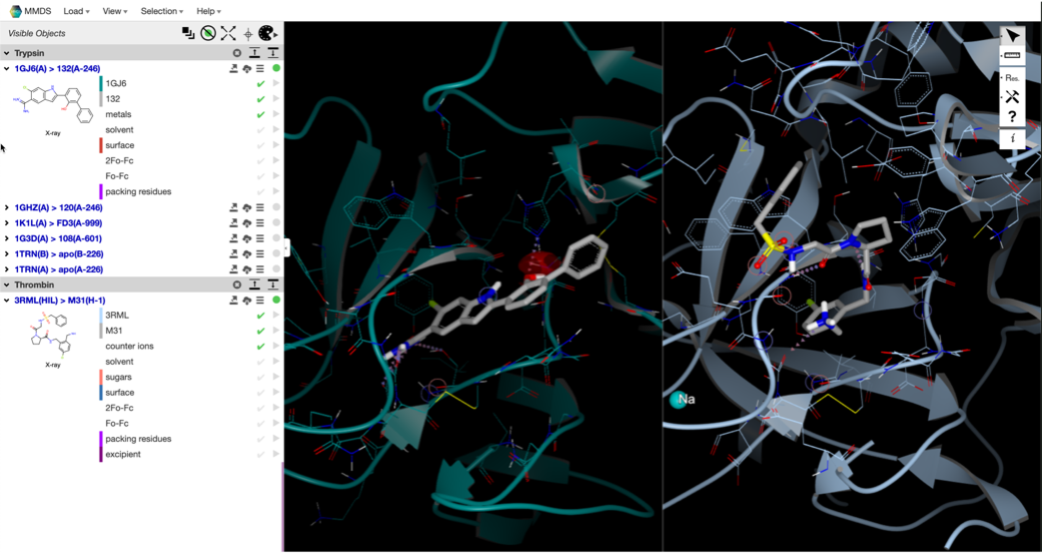
- Customizable 3D Viewing and Comparison
Customize your 3D-model view so you can work intuitively and quickly.
- Choose your model's style, coloring, labeling, and water & H-bond preferences.
- Select the mouse-mapping option that works best for you.
- Search for similar structures and superpose 3D models for quick comparison.
- Compare protein sequences and highlight backbone deviations.
* Available with the Orion’s optional Managed Stack platform choice.
References:
- Grebner C, Norrby M, Enström J, Nilsson I, Hogner A, Henriksson J, Westin J, Faramarzi F, Werner P, Boström J. 3D-Lab: a collaborative web-based platform for molecular modeling. Future Med Chem. 2016 Sep;8(14):1739-52. doi: 10.4155/fmc-2016-0081. Epub 2016 Aug 31. PMID: 27577860.
- IUPHAR/BPS Guide to Pharmacology
- Warren GL, Do TD, Kelley BP, Nicholls A, Warren SD. Essential considerations for using protein-ligand structures in drug discovery. Drug Discov Today. 2012 Dec;17(23-24):1270-81. doi: 10.1016/j.drudis.2012.06.011. Epub 2012 Jun 21. PMID: 22728777.
Webinar: OpenEye's Free energy prediction for drug discovery: Ideas at breakfast, discoveries by lunch
Science Brief: Binding Free Energy Redefined: Accurate, Fast, Affordable
CUP XXV - Santa Fe March 10-12, 2026
Webinar: Novel Hits from Beyond the Known: ROCS X
Resources
Glimpse the Future through News, Events, Webinars and more
News
ROCS X: AI-Enabled Molecular Search Unlocks Trillions
Webinar
Webinar: OpenEye's Free energy prediction for drug discovery: Ideas at breakfast, discoveries by lunch


