OpenEye Scientific is pleased to announce the release of OpenEye Applications v2020.0 and OpenEye Toolkits v2020.0, with several key updates and improved features to increase speed and functionality for molecular design. These packages include the usual support for Linux, macOS, and Windows. In addition, the toolkit libraries include the usual support for C++, C#, Java, and Python.
HIGHLIGHTS
- OpenEye Toolkits and Applications are now integrated into a single release cycle, simplifying customer updates
- GPU-OMEGA is now available for OMEGA (torsion driving mode) in both application and toolkit, resulting in a 4-30x speed increase
- Improved performance for FastROCS TK, which can now use both shape and color in overlay optimization
- Protein loop modeling is now available in the SPRUCE application and toolkit,
Spruce TK. This facility uses a template database that can be updated via a separate application - SZYBKI now provides custom force fields based on SMIRNOFF (SMIRKS Native Open Force Field), a small-molecule force field from the Open Force Field Initiative
- A preliminary API improving performance and memory management for OERecord and associated classes now is part of OEChem TK
OpenEye Toolkits and Applications Release Cycle
OpenEye Toolkits and Applications have been integrated into a single release cycle. This should simplify the maintenance of OpenEye software at customer sites. In addition, this integration reduces release process overhead, freeing developer time for new features and bug fixes.
GPU-OMEGA: GPU-Accelerated Torsion Driving
The 2020.0 release introduces GPU-OMEGA to provide accelerated torsion driving, particularly in the dense sampling mode. GPU-OMEGA is integrated into the existing OMEGA application and Omega TK; no additional user actions are required to make use of this new feature.
GPU-OMEGA detects any supported Nvidia GPU on Linux platforms and accelerates performance unless explicitly suppressed by using the option -useGPU false in OMEGA or through the option OETorDriveOptions.SetUseGPU in Omega TK. GPU-OMEGA is available with all sampling modes except macrocycle, which does not use the torsion driving method. For more details on GPU-OMEGA, including prerequisites and additional requirements for using dense mode, please see GPU-OMEGA.
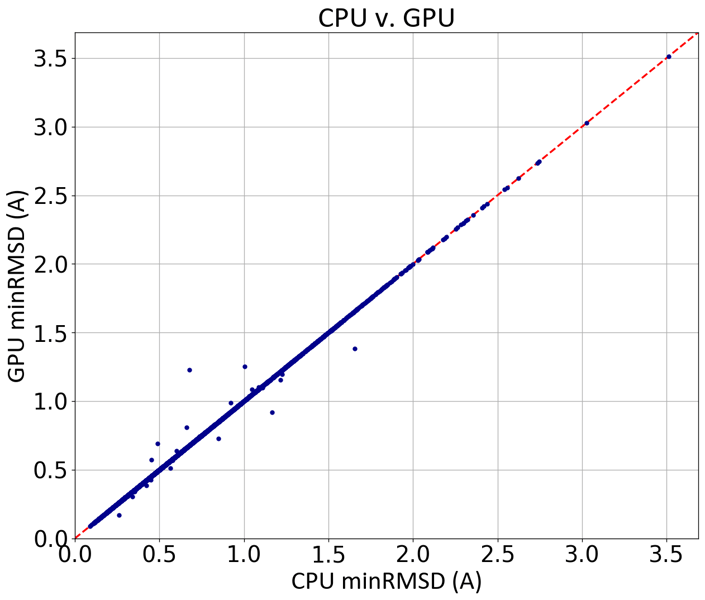
GPU-OMEGA performs at the same high level of accuracy as Omega TK, as shown below using a filtered subset of the Platinum dataset [Friedrich-2017] [Hawkins-2020]:
Comparison of the minimum RMSDs of CPU- and GPU-generated conformers against the experimental coordinates of a 3600-molecule subset of the Platinum dataset.
GPU-OMEGA was benchmarked against the CPU version on Amazon Web Services (P3 instance, with a single Nvidia Tesla V100 GPU) on all the available sampling modes (classic, dense, pose, rocs, and fastrocs) using a subset of ~6000 molecules from the GSK TCAMs dataset [Gamo-2010]. The most impressive speed-up is seen with the dense sampling mode, with a reduction in time from 120 hours to less than 4 hours.
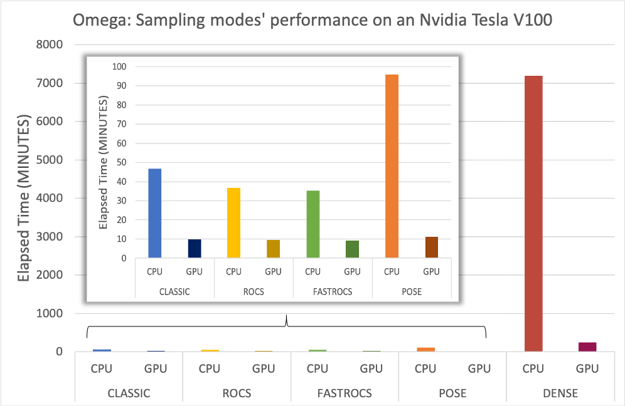
Elapsed time comparison between OMEGA or Omega TK and GPU-Omega on an AWS P3 instance when sampling a subset of the GSK-TCAMS dataset with classic, rocs, fastrocs, pose, and dense modes.
FastROCS TK: Color Optimization On the GPU
Overlay optimization by color, in addition to shape, is now available in FastROCS TK. Color adds chemistry representation to the superposition and similarity analysis process, facilitating identification of compounds that are similar in both shape and chemistry.
FastROCS TK with color optimization was carried out on single AWS GPU instances with NVIDIA Tesla K80, NVIDIA Tesla M60, and NVIDIA Tesla V100 GPUs and compared against those without color optimization:
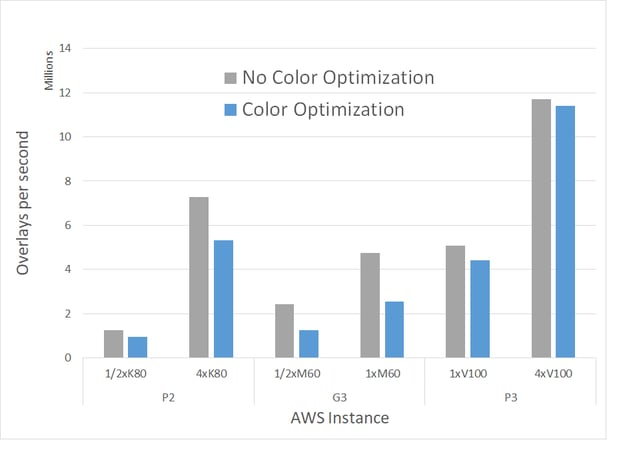
Performance comparison for FastROCS TK with and without color optimization in the 2020.0 release, using OpenEye’s standard benchmark dataset of 14M conformers on AWS.
Previous versions of FastROCS TK optimized the shape score between two molecules, followed by a single point calculation for color. This approach, while fast, can underestimate color similarity. An example of an improvement when optimizing both color and shape overlap rather than just shape overlap is shown below for XIAP-BIR3 [Ndubaku-2009]:
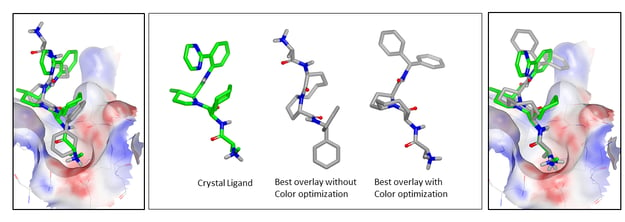
Example of pose improvement with color optimization in FastROCS TK.
On the left is the XIAP-BIR3 binding site showing the best overlay without color optimization of a known active ligand (from the DUDE dataset [Mysinger-2012]) to the crystallographic ligand in green.
The center images show a known active aligned to the crystallographic ligand by FastROCS TK without and with color optimization, respectively. Without color optimization, the known active shows relatively low color similarity. When color optimization is used the color similarity and “combo” scores are substantially better, and as might be expected given the substantial chemical similarity.
On the right is the overlay in the binding site of XIAP-BIR3, occupying a similar pose to the crystallographic ligand.
SPRUCE: Protein Loop Modeling
The 2020.0 release adds protein loop modeling to the SPRUCE application and Spruce TK.
This approach relies on a template database derived from structures in the public RCSB Protein Data Bank. The database, which has been built for modeling loops that are 4-20 residues long by default, is available for download in a platform-independent format.
A separate application, LoopDB_Builder, can append additional loop data to the existing database, e.g. from in-house structures, or completely rebuild the database with different specifications.
This approach has been validated on a public dataset from Rossi et al. [Rossi-2007]. The results shown below were generated by building the excised loops back into the dataset, where the specific structures themselves were not part of the template database, and used the same success criterion as in the paper (<2.5Å backbone RMSD).
The results are generally good with a median RMSD of 0.6-0.7Å (blue dots), irrespective of the length of the modeled loop. The mean RMSD (green dots) is biased higher as some unsuccessful cases (gray bar) can carry “outlier” weight. Only in one case (out of ~200 loops), was a result not produced (red bar), i.e., did not survive the search and filtering steps.
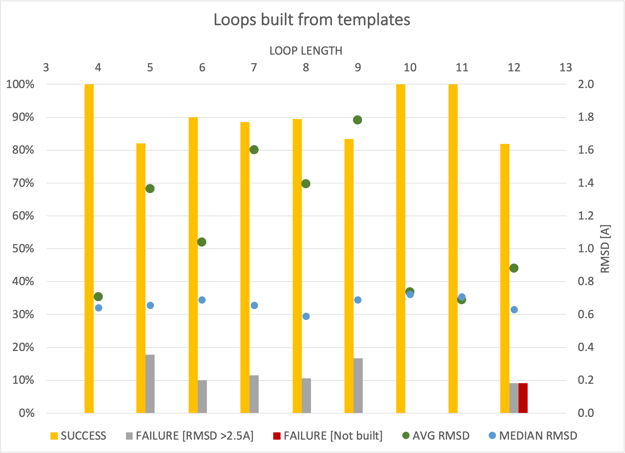
Performance assessment on 203 loops ranging from 4-12 residues in length.
SZYBKI: Improving Forcefield Accuracy and Coverage
SZYBKI now has a new forcefield for small molecule energetics, Parsley_OpenFF1.1.1 from the Open Force Field Initiative.
SZYBKI and FreeForm can now use custom force fields based on SMIRNOFF (SMIRKS Native Open Force Field).
The -ff parameter in both SZYBKI and FreeForm now accepts XML files with SMIRNOFF specifications in addition to the predefined names for built-in force fields (such as MMFF94 and Parsley). In Szybki TK, support for custom force fields is provided through two new methods, OESzybkiGeneralOptions.SetForceField and OESzybkiGeneralOptions.GetForceField.
OEChem TK: Data Record Preliminary API
A preliminary API for OERecord and its associated classes is now part of OEChem TK.
OERecord is a data container for storing and transmitting strongly typed data and its associated metadata. A pure Python implementation of OERecord has been in use by Orion and Floe; the new C++ implementation improves performance and memory management, and makes data records available to OpenEye toolkit code. The API is only available in C++ and Python at this time.
GENERAL NOTICES
Applications
- All applications, including GUI applications, are now available in the RHEL8 application bundle except for GPU-OMEGA, the CUDA-enabled feature of OMEGA. GPU-OMEGA will be supported in the next release when the toolkits have been upgraded to CUDA 10.
- All applications were built using OEToolkits 2020.0. The previous versions were built using OEToolkits 2019.Oct.
Toolkits
- Memory management in the Java toolkits no longer relies on the JVM to clean up objects. This significantly improves Java toolkit performance but requires users to manage memory. See Best Practices for Java for current best practices.
- C++ toolkits are now available for GCC 8.x
- Visual Studio 2019 toolkits are now available for C++ and C#
PLATFORM SUPPORT
Applications
- The application bundle for macOS is now notarized and can be run on macOS Catalina 10.15. Apple notarization gives users of our software more confidence that the Developer ID-signed software has been checked by Apple for malicious components.
- This is the last release to support Ubuntu16.04. Support for Ubuntu20.04 will be added in the next release.
- macOS 10.12 is no longer supported.
Toolkits
- Support for macOS Catalina 10.15 has been added. macOS 10.12 is no longer supported.
- RHEL8 is supported across all toolkits except for the CUDA-enabled toolkits and features. FastROCS TK, GPU-OMEGA, and GraphSim TK’s CUDA fast fingerprints are not supported. Support for CUDA-enabled features will be added in the next release when the toolkits have been upgraded to CUDA 10.
- Support for Python 3.8 has been added for all supported platforms. Python 3.6 is no longer supported for Windows toolkits and Python 3.5 is no longer supported for macOS and Linux.
- Java toolkits are now available for macOS.
- This is the last release to support Ubuntu16.04. Support for Ubuntu20.04 will be added in the next release.
See Platform Support for full and specific details.
AVAILABILITY
OpenEye Applications v2020.0 and OpenEye Toolkits v2020.0 are now available for download. Existing licenses will continue to work, but if a new license is required, please contact your account manager or email sales@eyesopen.com.
Please see the application and toolkit release notes for full and specific details on improvements and fixes.
RESOURCES
|
About OpenEye Scientific
OpenEye Scientific is an industry leader in computational molecular design based on decades of delivering rapid, robust, and scalable software, toolkits, and technology and design services across the drug discovery process. Our scientific approach has focused on visualizing and analyzing the shape and electrostatics of molecular 3D structures to inform and guide the advancement of pharmaceuticals and biotechnology. OpenEye recently integrated its applications and toolkits into Orion, the only cloud-native, fully integrated molecular design platform. Combining unlimited computation and storage with powerful tools for data sharing, visualization and analysis in an open development platform, Orion offers unprecedented capabilities for drug discovery and optimization. Founded in 1997, OpenEye is a privately held company headquartered in Santa Fe, N.M., with offices in Boston, Mass.; Cologne, Germany; and Tokyo, Japan.