
OMEGA
OMEGA is designed to sample accurately the conformational space of a wide variety of molecules. It generates multi-conformer databases with high speed and reliability, using two different algorithms, one for broadly drug-like molecules and one for highly flexible linear or macrocyclic molecules.
OMEGA samples the conformational space of drug-like molecules at speeds of hundreds of thousands of compounds per day on a CPU, and over a million molecules per day on a GPU.
OMEGA is very effective at reproducing bioactive conformations [1,2], and provides an optimal balance between speed and performance when used on large compound databases [3].
OMEGA conformational databases can be used as input to a variety of applications including docking engines (FRED), shape comparison tools (ROCS®), ligand posing (POSIT), and pharmacophore perception algorithms.
OMEGA produces high-quality ensembles of conformations, with higher accuracy than those from many other methods [4]. It has also been found to be the fastest of all commercially available conformer generators for drug-like molecules [5] and one of the fastest for macrocycles [6]. For more detailed information on OMEGA, check out the link below:
Documentation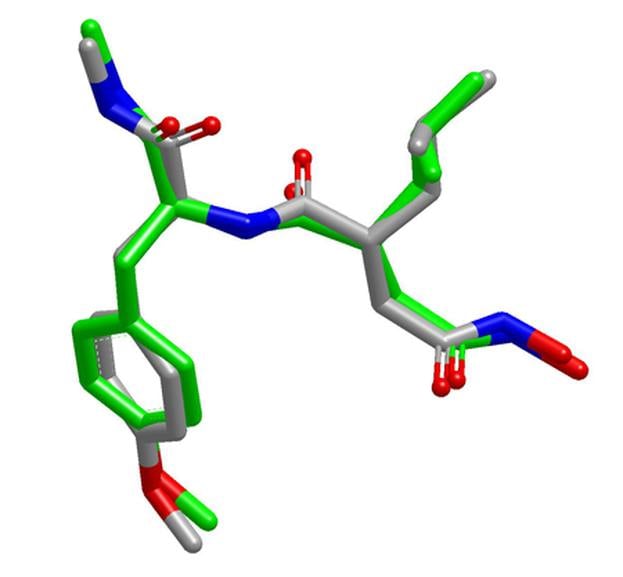
Features
- Very rapid (<0.3 sec/molecule), systematic and rule-based conformer search
- Converts from 1D or 2D to 3D using distance bounds methods
- Diverse ensemble selection based on RMS deviation and strain energy
- User-configurable search resolution
- Automatic superposition of structural features
- Excellent reproduction of solid-state and solution conformations of drug-like molecules and macrocycles
- Distributed processing via MPI for all supported platforms
Learn more
OpenEye's OMEGA produces high-quality ensembles of bioactive conformations with the fastest speed when compared to other commercially available conformer generators for drug-like molecule.
For science details, WATCH OpenEye’s miniWebinar recording from August 2023 on OMEGA by Paul Hawkins, Ph.D., Product Evangelist, OpenEye, Cadence Molecular Sciences.
References
- Conformer Generation with OMEGA: Algorithm and Validation Using High Quality Structures from the Protein Databank and Cambridge Structural Database, Hawkins, P.C.D., Skillman, A.G., Warren, G.L. Ellingson, B.A. and Stahl, M.T., J. Chem. Inf. Model., 2010, 50, 572-584.
- Conformer generation with OMEGA: Learning from the dataset and analysis of failures, Hawkins, P.C.D., Nicholls, A.N., J. Chem. Inf. Model., 2012, 52, 2919-2936.
- Conformational Analysis of Drug-like Molecules Bound to Proteins: An Extensive Study of Ligand Reorganization upon Binding, Perola, E. and Charifson, P.S., J. Med. Chem. 2004,47, 2499-2510.
- High-Quality Dataset of Protein-Bound Ligand Conformations and its Application to Benchmarking Conformer Ensemble Generators, Friedrich, N.-O., Meyder, A., de Bruyn Kops, C., Sommer, K., Fachsenberg, F., Rarey, M., Kirchmair, J. J. Chem. Inf. Model. 2017, 57, 529-539.
- Benchmarking Conformer Ensemble Generators, Friedrich, N.-O. de Bruyn Kops, C. Fachsenberg, F. Sommer, K., Rarey, M. Kirchmair, J. J. Chem. Inf. Model. 2017, 57, 2719-2728.
- Decisions with Confidence: Application to the Conformation Sampling of Molecules in the Solid State, Hawkins, P.C.D., Wlodek, S., J. Chem. Inf. Model., 2020, 60, 3518-3533. (doi: 10.1021/acs.jcim.0c00358)
Over 800x Speed-Up on Orion to Enable Rapid and Reliable Drug Design
Expanding Orion’s Capabilities with AI
miniWebinar | Shape and color: A unifying principle for modeling molecules
miniWebinar: Faster, Larger, Smarter: Filling the Funnel for Ultra-Large Scale Virtual Screening
Resources
Glimpse the Future through News, Events, Webinars and more
Webinar
miniWebinar: Opening up the druggable universe one cryptic pocket at a time
News
Over 800x Speed-Up on Orion to Enable Rapid and Reliable Drug Design


