
Trusted Science. Delivered the Way You Need.
AbXtract™ Module for Sequence Analysis
Analyze your Next Generation Sequencing (NGS) and Sanger data on the cloud with the Orion® molecular design platform. These capabilities are part of AbXtract™, a module within Orion's Antibody Discovery Suite. AbXtract was developed in partnership with Specifica, founded by Andrew Bradbury, PhD, an innovator in novel antibody technologies. Contact us for a demo on how AbXtract can help you obtain potent leads with the most favorable developability and biophysical profiles.
Compared to conventional colony screening methods, NGS screening with AbXtract helps teams:
- Uncover more leads. Increase the number of clonotype leads five- to ten-fold compared to random colony screening
- Increase sequence diversity and cluster representation. Explore the entire sequence diversity within selected populations, even rare clones that are typically missed by low-throughput methods that favor more abundant antibodies
- Prioritize promising leads. Tie in known data, and even low-throughput assay data, to prioritize leads with the most favorable developability and biophysical profiles
- Minimize costs. Achieve high throughput for a fraction of the cost of conventional assay runs
- Equip the entire discovery team. Automated workflows for novice users, while allowing expert users to fully configure their settings
Antibody discovery projects that were previously out of reach because of software, hardware, computation, or data sharing limitations have now become possible with AbXtract on Orion. -- Andrew Bradbury, PhD, Specifica
From Millions of Sequences to a Select Few
AbXtract has the cloud-based compute power to process tens of millions of sequences into meaningful information. Its sophisticated machine-learning algorithms and workflows help antibody engineers and bioinformaticians parse through this vast amount of data to characterize sequences and extract sequence- and functional-based features. Once promising leads are identified, visual models can further aid in decision-making.
User Friendly
Get Leads from Your Data in Three Guided Steps
-
- Upload sequence files into Orion
- NGS: FASTQ, FASTA
- Low-throughput: FASTA, FASTQ, EXCEL, TSV, CSV
- Perform FASTQ quality filtering
- Perform simple demultiplexing of highly multiplexed experiments
- Annotate to identify CDRs and framework regions of interest (IMGT, Kabat, Chothia, or custom annotation)
- Produce annotated records ideally suited for antibody discovery
- Upload sequence files into Orion
-
- Extract features
- Avoid contaminants
- Quantify population statistics
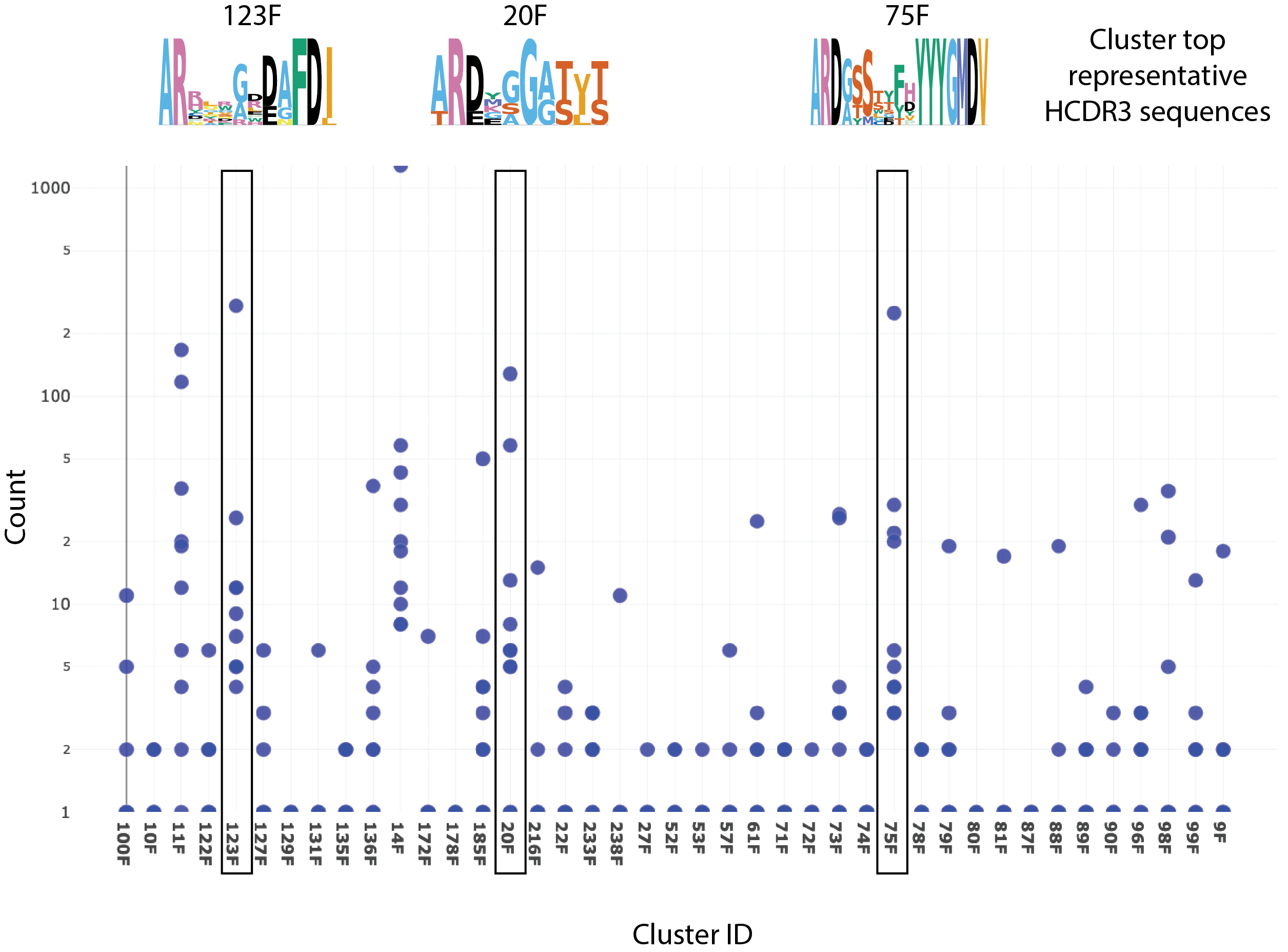
- Maximize CDR diversity
- Classify functionality
- Obtain frequency of clones by population
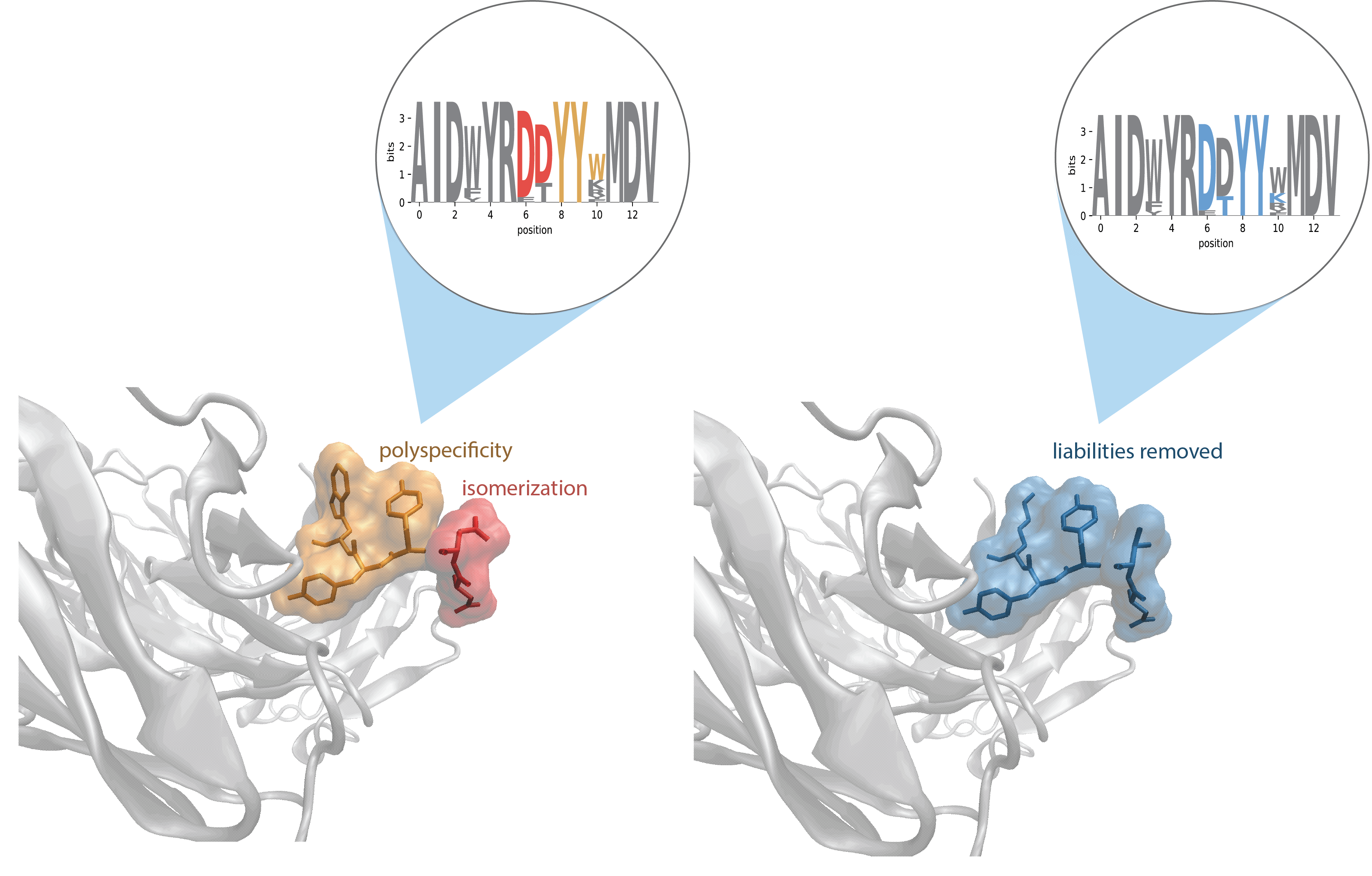
- Eliminate bad sequences (e.g., those seen before or with known biophysical liabilities)
- Assess the scaffold distribution
- Identify relationships between and within sequence clusters to avoid redundancy
- Leverage known data for various sequence populations under different experimental conditions
- Look at round-to-round enrichment to identify true binders and better-affinity binders
- Couple in low-throughput screening-assay data for sequences with desired features
- Identify overlap among different target populations
-
- Identify best performers using NGS metrics derived from experimentally validated studies
- Leverage prior knowledge to:
- Maximize diversity
- Reduce risk
- Minimize redundancy
- Optimize time to pre-clinical trials
- Maximize the selection of leads with very distinct sequence-based properties
- Perform unsupervised physicochemical clustering to identify clusters sharing similar binding modes
- Easily select unexplored antibodies with likely distinct paratope space
- Identify additional clones that share the same features as those characterized, such as:
- Antibodies from the same cluster lacking specific sequence liabilities
- Antibodies from the same cluster that span a broad affinity range
- Quantify developmental liabilities from common and customized references
- Perform region of interest (ROI)-based enrichment calculation across selection rounds / populations
- Identify population overlap at deeper sequencing depth.
- Identify the relative abundance of clones that bind distinct targets or variants
- Identify NGS clones that map to already characterized populations
Learn More
Orion’s Antibody Discovery Suite helps you get better leads from your NGS sequence data.
WATCH OpenEye’s miniWebinar recording "Maximizing the Panel of Diverse Leads from a Wide Array of Antibodies Recognizing the Same Epitope with AbXtract™" by M. Frank Erasmus, MS, PhD, Head of Bioinformatics at Specifica, from Thursday, August 25, 2022.
WATCH OpenEye’s miniWebinar recording "How AbXtract™ and Deeper Sequencing can Guide your Antibody Lead Selection and Prioritization" by M. Frank Erasmus, MS, PhD, Head of Bioinformatics at Specifica, from June 16, 2022.
Additional Resources
- Ferrara, F, Erasmus, M.F., D’Angelo, S., Leal-Lopes, C., Teixeira, A., Choudhary, A., Honnen, W., Calianese, D., Huang, D., Burton, D., Pinter, A., Bradbury, A.R.M. A pandemic-enabled comparison of antibody discovery platforms: a naïve library directly delivering antibodies as potent as immune sources. Nature Comm. 13:462 (2022).
- Teixeira, A., Erasmus, M.F., D’Angelo, S., Naranjo, L., Ferrara, F., Leal-Lopes, CC., Durrant, O., Galmiche, C., Morelli, A., Scott-Tucker, A., and Bradbury, A.R.M. Drug-like antibodies with high affinity, diversity and developability straight from next-generation antibody libraries. MAbs vol 13, 1 (2021).
- Erasmus, M.F., D’Angelo, S., Ferrara, F., Naranjo, L., Teixeira, A., Buonpane, R., Stewart, S.M., Nastri, H.G., and Bradbury, A.R.M. A single donor is sufficient to produce a highly functional in vitro antibody library. Commun Biol 4, 350 (2021).
- Bradbury, A. R. M., Erasmus, M. F. & Teixeira, A. Antibody libraries with maximized antibody developability characteristics. US patent (2020).
miniCUP Cambridge 2025
Cadence Announces New Life Sciences Leadership
Expanding Orion’s Capabilities with AI
miniWebinar: Faster, Larger, Smarter: Filling the Funnel for Ultra-Large Scale Virtual Screening
Resources
Glimpse the Future through News, Events, Webinars and more
Event
miniCUP Basel 2025
Event
miniCUP Cambridge 2025


