
SiteHopper
Comparing protein binding sites is an important method in drug discovery that plays a part in a wide variety of structure-centric pursuits. One challenge for binding site comparison methods is the broad goals of such approaches: from identifying subtle differences between largely similar sites in induced fit analysis or understanding selectivity, to identifying general similarity for drug repurposing or target fishing searches.
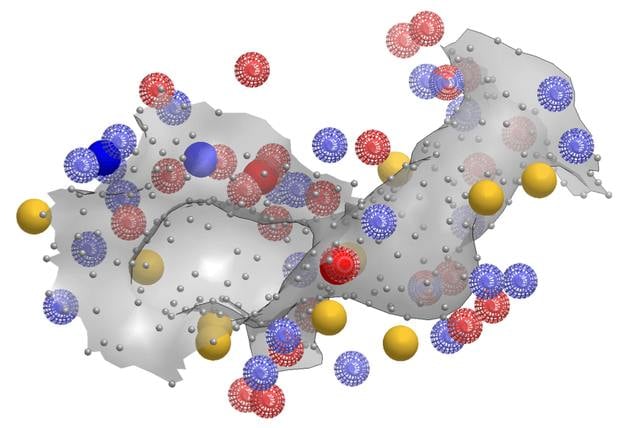
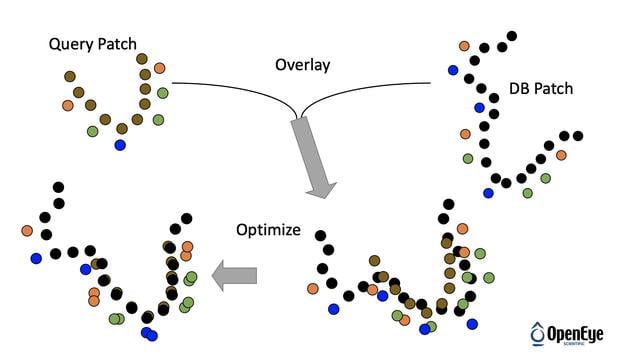
SiteHopper builds on the industry best-in-class 3D shape and chemical feature ligand comparison application (ROCS®) from OpenEye to the domain of protein comparison. Treating binding sites as constructs with a defined shape and set of chemical features allows for rapid searching of a database of known protein structures. You can generate your own protein database to search, or use OpenEye's ready to use protein-ligand binding sites database for your convenience.
What is available in the prepared SiteHopper database?
Ready to search SiteHopper patch databases are available; over 200K known protein-ligand binding sites, and over 2 Million potential protein binding sites. The datasets were generated from available PDB files and AlphaFold2 structures.
Can I run SiteHopper on GPU or Cloud?
You can compare a query protein to a database of hundreds of thousands (or millions) of protein binding sites efficiently, thanks to a multi-threaded searching algorithm (GPU on Linux) or harness the scalability of cloud computing.
How can SiteHopper helps in my early drug discovery?
The speed and flexibility of SiteHopper opens new avenues in drug repurposing, predicting off target effects, and polypharmacology among other uses in drug design projects.
For more detailed information on SiteHopper, check out the link below:
References
- Prediction of Protein Pairs Sharing Common Active Ligands Using Protein Sequence, Structure, and Ligand Similarity. Yu-Chen Chen, Robert Tolbert, Alex M. Aronov, Georgia McGaughey, W. Patrick Walters, and Lidio Meirele. Journal of Chemical Information and Modeling 2016 56 (9), 1734-1745. DOI: 10.1021/acs.jcim.6b00118
- Privileged Structures and Polypharmacology within and between Protein Families. Joshua Meyers, Nicola E. A. Chessum, Salyha Ali, N. Yi Mok, Birgit Wilding, A. Elisa Pasqua, Martin Rowlands, Michael J. Tucker, Lindsay E. Evans, Carl S. Rye, Lisa O’Fee, Yann-Vaï Le Bihan, Rosemary Burke, Michael Carter, Paul Workman, Julian Blagg, Nathan Brown, Rob L. M. van Montfort, Keith Jones, and Matthew D. Cheeseman. ACS Medicinal Chemistry Letters 2018 9 (12), 1199-1204. DOI: 10.1021/acsmedchemlett.8b00364
- SiteHopper - a unique tool for binding site comparison. Batista, J., Hawkins, P.C., Tolbert, R. et al. SiteHopper - a unique tool for binding site comparison. J Cheminform 6, P57 (2014). https://doi.org/10.1186/1758-2946-6-S1-P57
Webinar: OpenEye's Free energy prediction for drug discovery: Ideas at breakfast, discoveries by lunch
Science Brief: Binding Free Energy Redefined: Accurate, Fast, Affordable
Webinar: ML-Enabled integration of affinity prediction and lead discovery: 3D-QSAR
CUP XXV - Santa Fe March 10-12, 2026
Resources
Glimpse the Future through News, Events, Webinars and more
News
ROCS X: AI-Enabled Molecular Search Unlocks Trillions
Webinar
Webinar: OpenEye's Free energy prediction for drug discovery: Ideas at breakfast, discoveries by lunch


